Software
This is a list of software programs that I developed.
Variant Calling and Integration (Long-read DNA and RNA)
Exacto (EXacto Accurate Characterization of Transcriptomes and genOmes) is a Python package with a high-performance Rust backend for comprehensive variant identification and analysis using long-read sequencing data. It identifies both somatic and germline DNA variants as well as RNA variants, and integrates them to predict mutant proteoforms using full-length RNA sequences for neoantigen discovery.
Bioinformatics Workflow Manager
Nexus (NEXtflow’s Ultimate Streamliner) is a Python package that runs unit-tested Nextflow workflows for various tasks in bioinformatics including alignment, assembly, variant calling, variant phasing, HLA typing, and RNA quantification. Every workflow runs on Docker containers for portability and reproducility.
Immunoassay Experiment Design and Deconvolution

ACE (ACE Configurator for ELISpot) is a standalone graphical-user-interface (GUI) software program that configures high-throughput (pooled) ELISpot experiments by a fine-tuned ESM-2 model that groups immunologically similar peptide sequences. It provides bench-ready spreadsheets of peptide-pool assignments for various well plate sizes (24, 96, and 384 wells). ACE also deconvolves pool spot counts to estimate peptide-level spot count and identifies hit peptides. The software is available on Windows 11, Mac, and Linux.
Mutational Signature Analysis
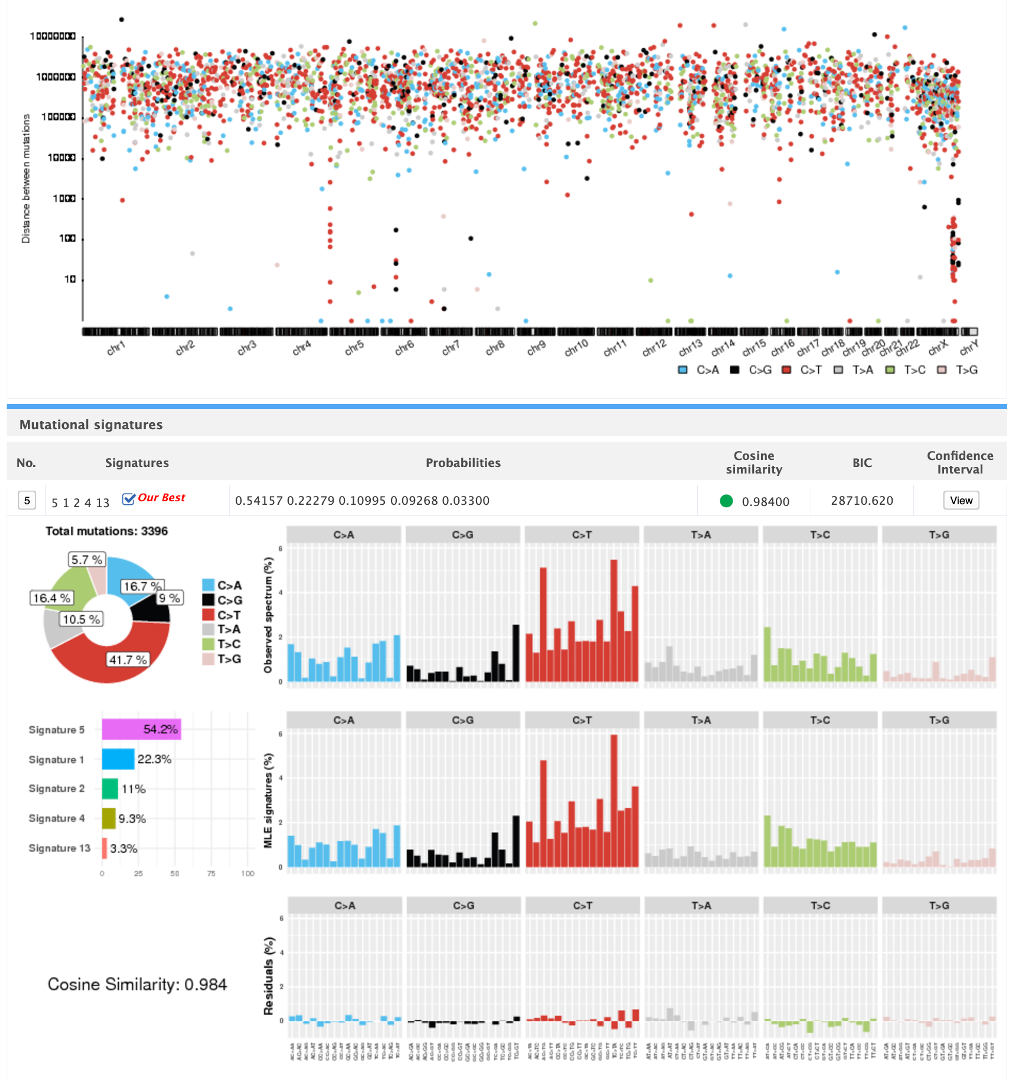
Mutalisk is a web-server that supports identification of COSMIC/PCAWG mutational signatures operative in a DNA sample.
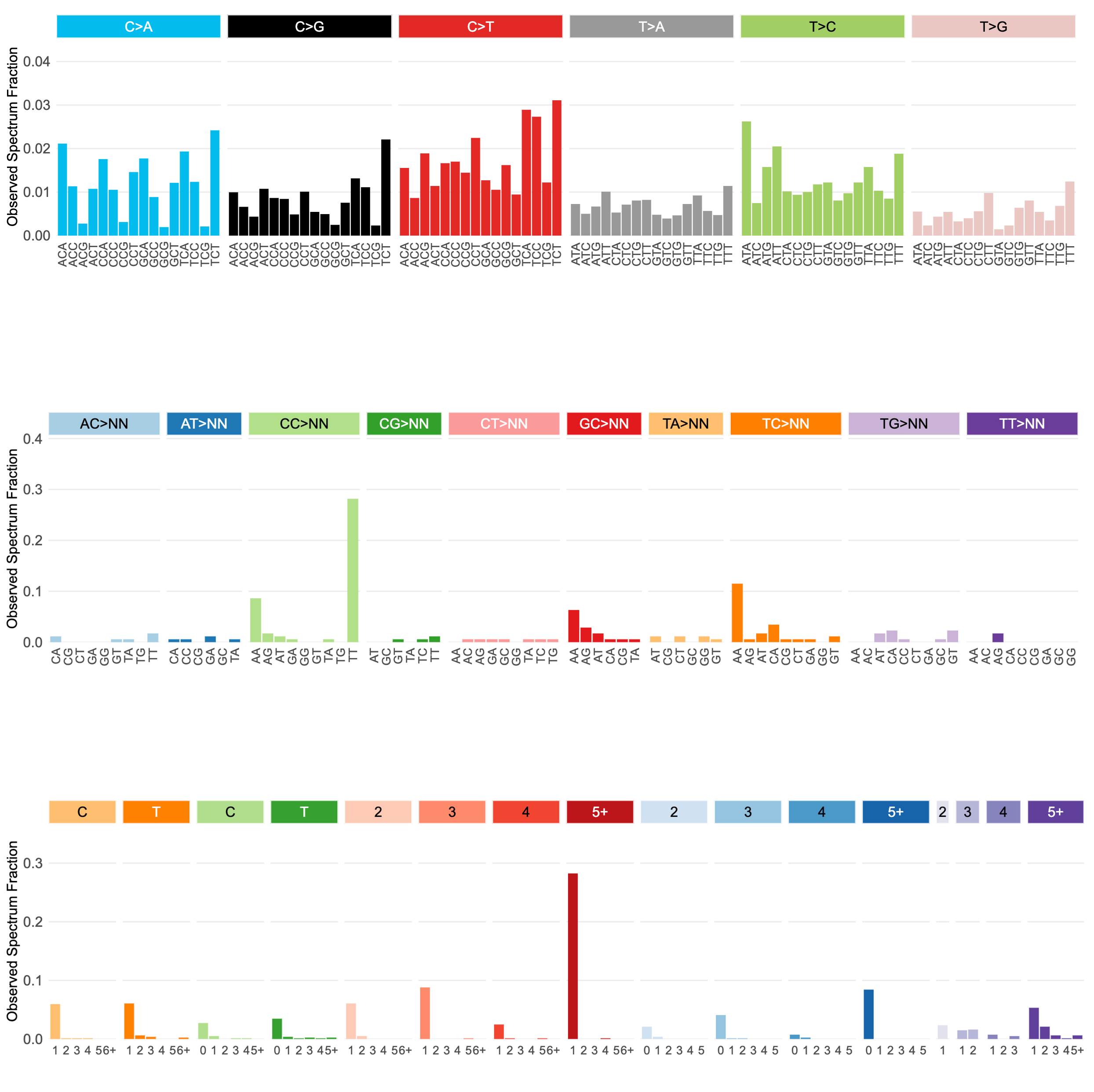
MutaliskR is an R package version of Mutalisk. It supports single-base substitution (SBS), double-base substitution (DBS) as well as small insertion and deletion (INDEL) signature identification. It also supports multiprocessing for scalability.
Variant Refinement
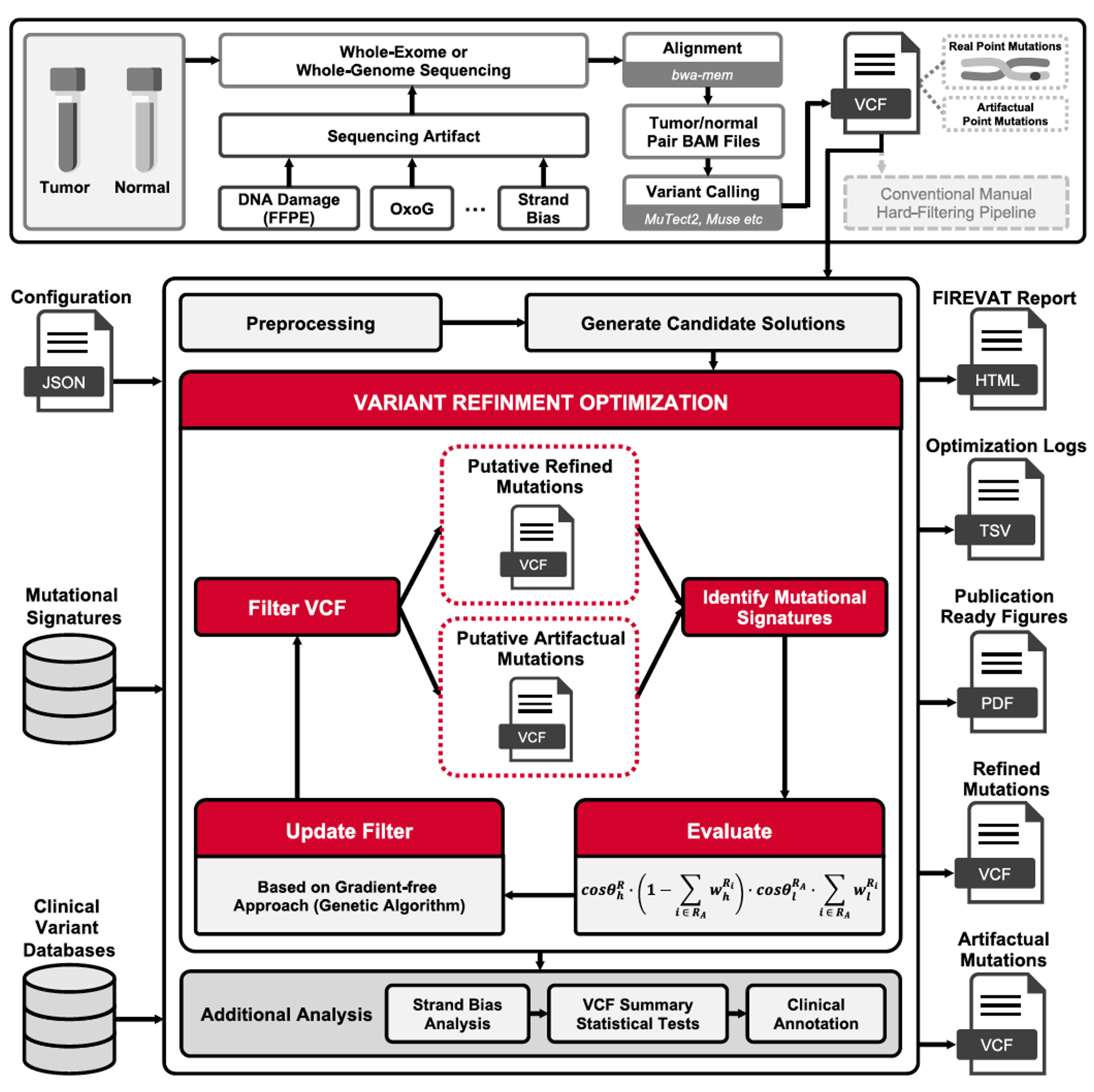
FIREVAT (FInding REliable Variants without ArTifacts) is a variant refinemnt tool that uses mutational signatures indicative of sequencing artifacts to filter out low-quality single-base DNA substitutions. FIREVAT generates an HTML report for convenient quality control.
Variant Standardization
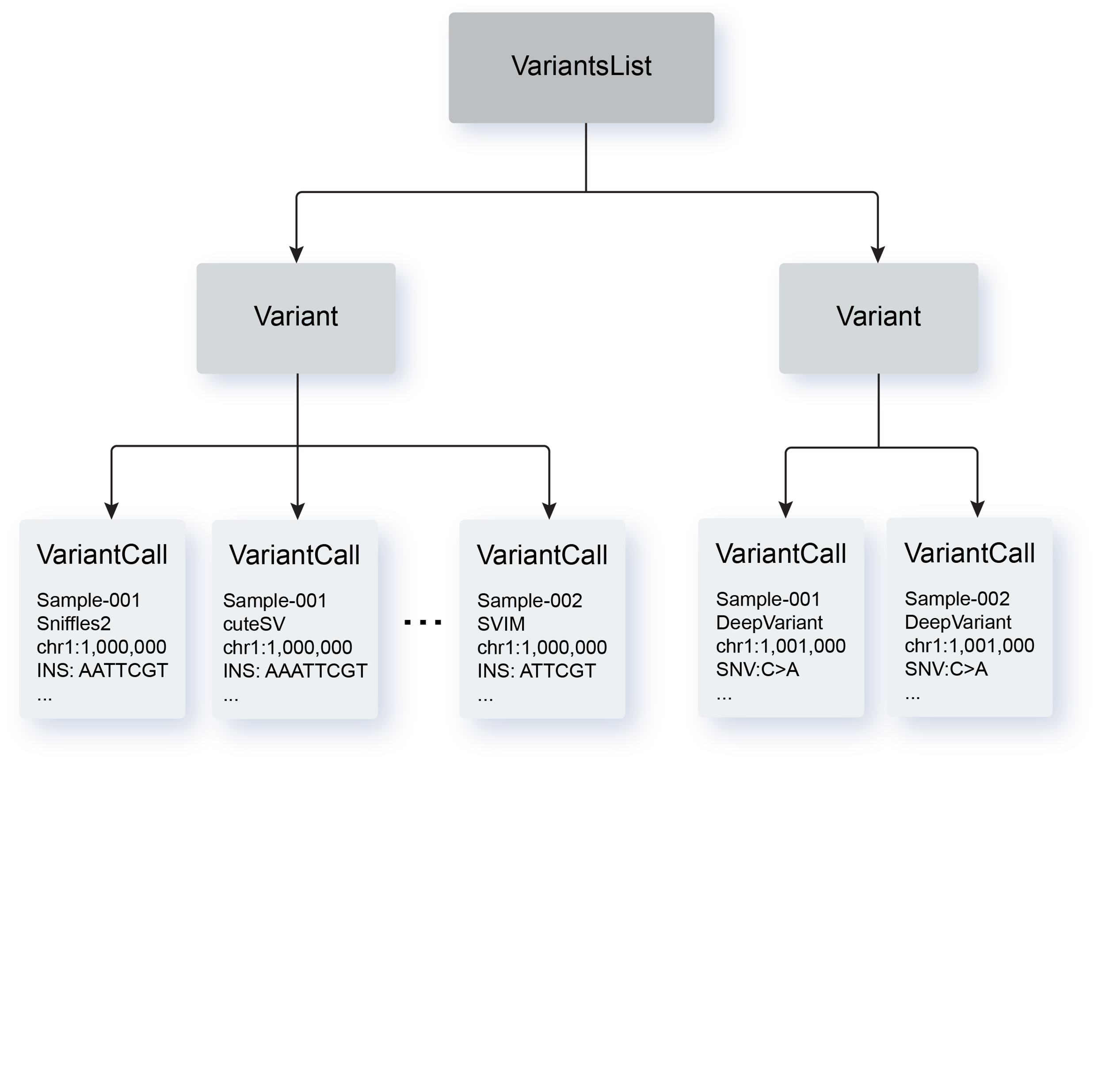
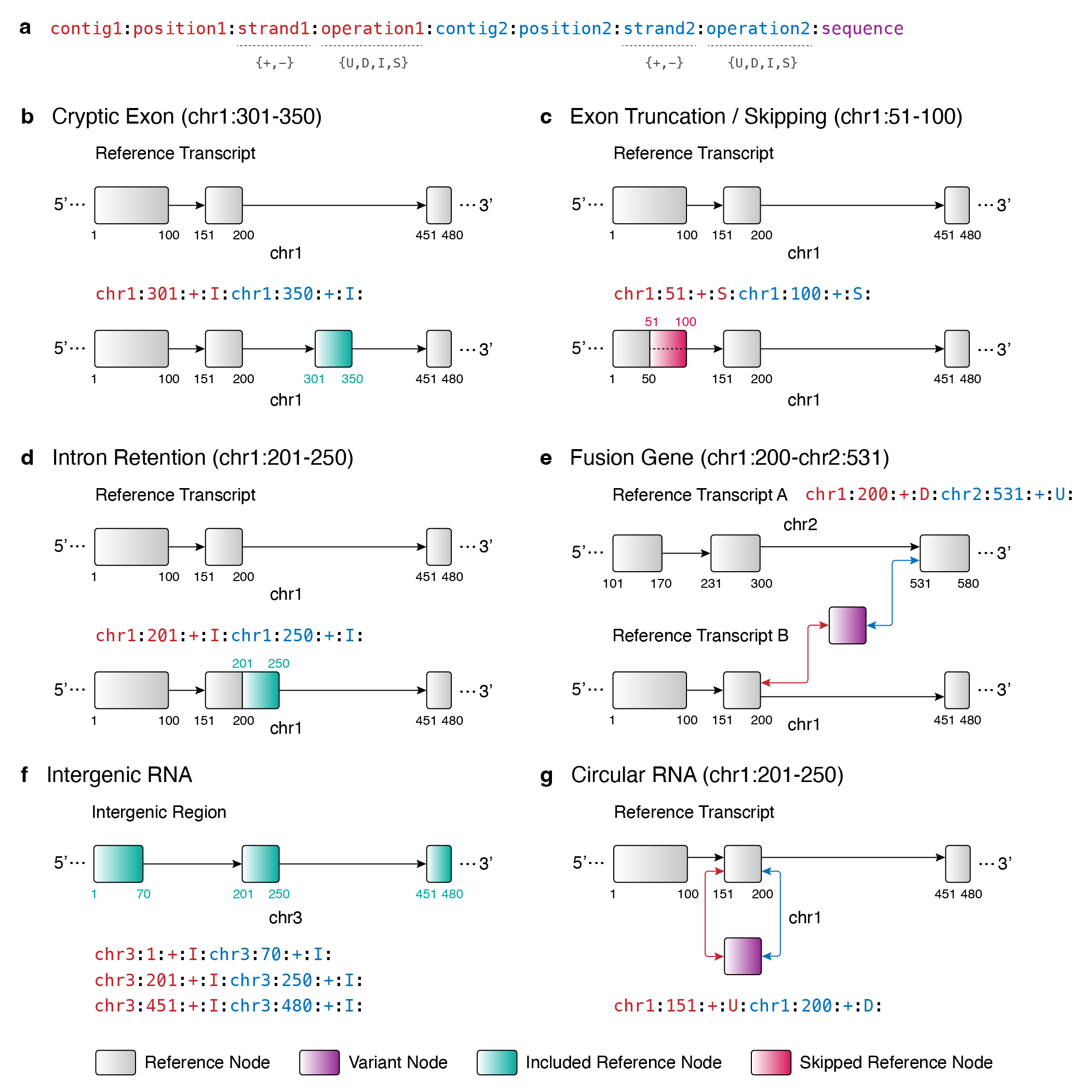
VSTOL (Variant Standardization, Tabulation, and Operations Library) is a Python package with Rust implementation that facilitates variant standardization of VCF files from various variant callers to a standardized grammar based on simplified sequence operations.